Human data from stages CS12 to CS23 can be submitted to the HUDSEN Gene Expression Database by e-mailing or posting images of experimental data (in situ hybridisation or immunohistochemistry) to this address.
The information we require is as follows:
Data can be entered into the database in two ways, either spatially mapped into a 3D context, or as a series of unmapped 2D images.
The data will be spatially mapped and entered into a local database. We will contact you once it is finished before uploading the data to the publicly accessible database.
It is very important that the sections/images being used to map data are of sufficient quality to allow the extraction of the data to be reliable. The image should be of sufficient size, contrast and brightness to see the expression clearly. The sections should also be of high enough quality to reduce folds and tears which create artefacts when the expression is thresholded.
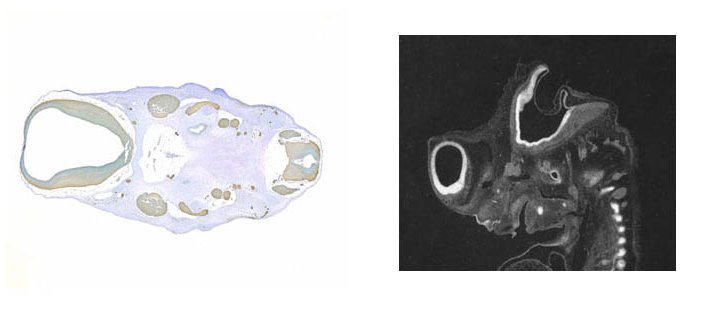
Good Quality
The sections below are two examples of the optimal data to be used for mapping purposes. The illumination is even and bright, the sections are completely intact while the slides are clearly devoid of debris and artefact. The staining is very strong and easily discernible from the background making the mapping process easier.
Unsuitable Section Data
The sections below are all examples data unsuitable for mapping (although they could be text annotated – see below). The section on the left is of a non-radioactive in situ experiment. The slide is very dirty with many artefacts, while the section itself has many folds and is very pale with little or no contrast between staining and background, making interpretation very difficult. The centre section, also non- radioactive in situ data, although clean, is over stained and the signal cannot easily be distinguished from the background. The radioactive in situ section on the right is folded over on itself and so will not be able to be mapped effectively. The folds in the tissue appear lighter than other areas and so may be incorrectly shown in the mapping process as signal data. The background is also quite dark which would make it harder to match sections to the 3D standard model.
Individual images or series of images can also be submitted to the database without being mapped into a 3D context. In this case the images should be of good enough quality, size, contrast and brightness to allow the expression to be distinguished easily. As there is no spatial information from the 3D model it is important to provide an annotated copy of the experimental section, or an adjacent histologically stained section, for orientation and comparison with the gene expression.